|
|
|
|
Fiber
Tracking
The
fiber tracking module in nordicICE implements the
feature of reconstructing axonal tracts in the central nervous system using
diffusion tensor imaging. The resulting three-dimensional architecture of the
white matter tracts can be studied using a designated 3D volume viewer. Currently,
nordicICE performs fiber tracking using the
algorithm commonly referred to as Fiber Assignment
by Continous Tracking (FACT) (Mori et.al Ann.
Neurol. 1999;45:265-269). Assuming that the
orientation of the largest component of the diagonalized diffusion tensor
represents the orientation of dominant axonal tracts, DTI provides a 3D
vectors field in which each vector represents the fiber orientation. The FACT
method basically consists of initiating a tract from a given seed-pixel
within such a vector field from which a line is propagated in both retrograde
and orthograde direction. The tracking is terminated when it reaches a pixel
with a fractional anisotropy index lower than a predefined threshold, or when
the inner product between two successive eigenvectors to be connected by the
tracking is smaller than a given threshold. NOTE:
The performance of the fiber tracking analysis is in
general highly dependent on both the quality of the input data
and the limitations of the implemented algorithm. The analysis may
fail to correctly reconstruct structures where diffusion pathways are overlapping
(crossing/kissing). Care should therefore be taken when interpreting the
results as the visualized fiber tracts do not necessarily correspond to real
physical structures. The
input to Fiber Tracking is the calculated diffusion tensors from a diffusion
weighted dataset. Thus prior to performing Fiber Tracking, you need to
perform the DTI analysis as outlined in DTI
analysis section . The fiber tracking visualization features are
accessed via the DTI interaction panel in the MPR viewer .
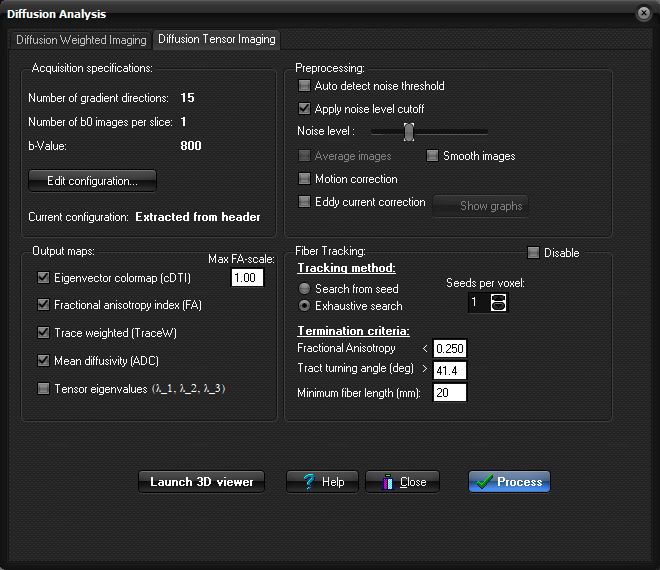
Tracking method
Tracking is always initiated from a set of
seed-pixels. These seed-pixels can either be defined as a subset of the image
volume ("Search from seed") or as the complete image volume
("Exhaustive search"). In the latter case, branching patterns are
more correctly reconstructed at the expense of processing time. Thus one of the two following methods must be selected in
the 'Settings' tabsheet: 1. Search from seed: Tracking is initiated from defined
seed-voxels. In this case, the seed-region has to be
defined within the image volume using the ROI feature (step 3). Optionally
two more ROIs can be defined requiring or excluding fiber passage in certain
regions of the brain. 2. Exhaustive search: Tracking is initiated from all
voxels within the volume, and tracts are followed until one of the
termination criteria are met. Optionally, up to 4 different ROIs can be
drawn within the image volume in order to select
only fibers that pass through the given regions of interest. Termination criteria
The line propagation must be terminated at some
point. The most intuitive termination criterion is the extent of anisotropy.
In a low anisotropy region, such as grey matter, there may not be a coherent
tract orientation within a pixel and the orientation of the largest principal
axis is more sensitive to noise errors. Another vital criterion is the angle
change between pixels. For the linear line propagation model being used,
large errors occur if the angle transition is large. It is therefore
preferable to set a threshold that prohibits a sharp turn during the
propagation. Fractional Anisotropy : If the Fractional Anisotropy index of the
voxel being propagated to is less than this threshold tracking is terminated.
Tract turning angle : If the angle (in degrees) between two successive
principal eigenvectors is larger than this threshold tracking is terminated. Number of voxels : Minimum number of assigned voxels required for
reconstructed tracks. This is typically used to filter out spurious tracks. Region(s)/Volume(s) of Interest
Up to 4 different VOI/ROIs can be
defined prior to performing fiber tracking. Each VOI/ROI can be assigned a
logical attribute - AND, OR, NOT - defining how the
fibers are selected:
See the DTI
interaction panel reference for a detailed description on how to use
these features.
|
|