|
|
Perfusion
Settings (options)
More
options relating to the perfusion analysis can be accessed from the <Options>
button at the bottom of the window.
The following menu
will appear:
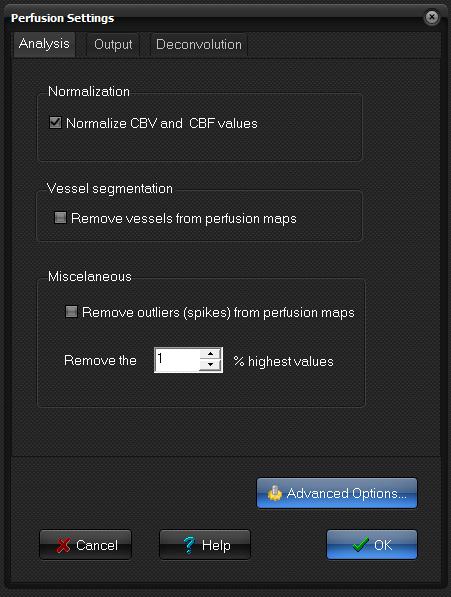
Analysis
- Normalization:
this will normalize the CBV and CBF maps to globally determined mean
value. The global mean value is calculated from all pixels determined to
represent 'normal' brain tissue (excluding noisy and otherwise
'abnormal' dynamic curves). See Emblem and Bjornerud
AJNR Am J Neuroradiol. 2009
Nov;30(10):1929-32. doi: 10.3174/ajnr.A1680 for more details on the method. This
normal brain reference mask can be either grey matter, white matter or both, and is selected in the Segmentation
tab of the Advanced Options menu. The reference mask segmentation is
automatically performed by the software. You can choose to generate the
segmented mask as an output map. This can be turned on in the Output tab
of the Options menu. Note that the MTT values will also be normalized
when this option is selected.
- Vessel segmentation: Automatic removal of vessel from perfusion maps.
The pixels classified as vessels are removed (pixel value set to zero)
from the output perfusion maps. The method is based on a cluster
analysis of the estimated perfusion related parameters to separate
vessels (both arteries and veins) from other tissues. For details
of the method used, see Emblem et al. Magn
Reson Med 2009 May;61(5):1210-7). NOTE: The vessel segmentation functionality
is meant as an aid in identifying vessels in perfusion maps and no
claims are made as to the accuracy of the method to truly identify
vessels.
- Miscellaneous: Remove outliers (spikes) from
perfusion maps: When gamma variate fitting is applied, or when the
input data is generally noise, the resulting perfusion values can, under
certain conditions, become artificially high if the fitting algorithm
fails resulting in 'spikes' in the perfusion maps. If this option is
selected nordicICE will attempt to detect outliers in the output pixel
values and remove these (i.e. by setting the
outlier pixels equal to the maximum of the 'normal' pixel range).
Output
Use
this menu to select which additional output maps you want to generate during
the perfusion analysis.
- Variance map:
The variance of time curve after baseline.
- Brain mask:
The segmented normal brain tissue, either white matter, grey matter or both, as set in Advanced Options -
Segmentation. Creates a binary image where each pixel is set to 1 or 0
depending on whether it is classified as normal tissue or not.
- Vessel mask: Creates
a binary image where each pixel is set to 1 or 0 depending on whether it
is classified as vessel or not.
- Mean baseline image
- Non-corrected CBV map: Option to create an additional CBV map where leakage
correction is not done (requires that the leakage correction option is
turned on).
- Non-segmented CBV map: Option to create an additional CBV map where vessel segmentation
is not done (requires that the vessel removal option is turned on).
- Chi-square map: This image represents the 'goodness of fit' of the
raw data to the gamma variate model function for each pixel (requires
that gamma variate fitting is done).
- Regularization index map: This image gives the regularization parameter in
the Tikhonov regularization (requires that Tikhonov regularization is
used, see advanced options for details).
Deconvolution
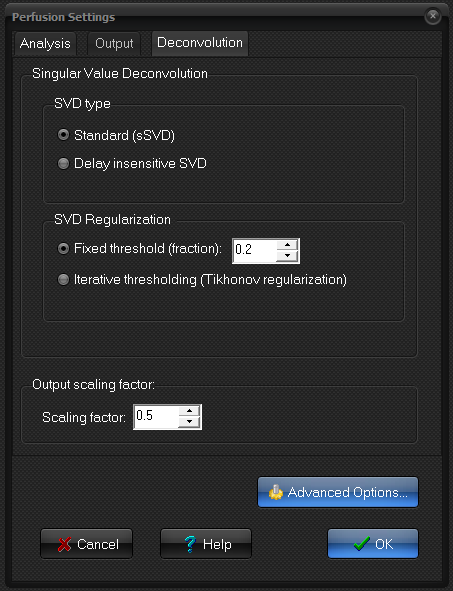
Singular Value Deconvolution
The
deconvolution procedure in nordicICE uses a mathematical technique called
singular value deconvolution (SVD), first suggested for use in MRI perfusion
analysis by Østergaard et al (Magn Reson Med 36:1996). One
critical requirement in SVD based deconvolution is
proper 'regularization' to obtain a stable solution and hence a robust
estimation of the perfusion related parameters. The SVD method requires a
threshold to be defined that specifies which of the components of the
decomposition process represent noise and hence should be eliminated from the
analysis. Different methods are available for determining the optimal
threshold. A low cutoff makes the solution more
sensitive to noise but gives more correct perfusion values in the noiseless
case. A large cutoff value provides a more robust
solution in the presence of noise but may also filter out relevant
information from the AIF response which can result in an over-estimation of
perfusion.
Two
SVD methods are included:
- Standard (sSVD).
- Delay insensitive SVD. To be used if AIF peak is
delayed in time compared to the tissue response.
Two
regularization methods are included:
- Fixed threshold: a set, fixed value is used. Components (of the
diagonal matrix from the SVD) less than
the specified fraction (relative to the largest value) are set to zero.
The threshold should be a fraction (of the maximum singular value)
between zero and one. The default value is 0.2. A larger value
will give more severe filtering and may result in under-estimation of perfusion,
whereas a too low value will introduce too much noise and resulting
spikes in the perfusion maps.
- Iterative using Tikonov
regularization. The
optimal filter threshold value is determined iteratively by finding the
'optimal' trade-off between a 'correct' solution and an oscillating
solution. For details of the method, see Hansen HC (SIAM
Journal on Scientific Computing 1993;14(6): 1487 – 1503). The number of
iterations to run can be specified in Advanced
perfusion settings. More iterations increase the processing
time.
See
Bjornerud and Emblem J Cereb
Blood Flow Metab. 2010 May;30(5):1066-78. doi: 10.1038/jcbfm.2010.4 for details.
Output scaling factor
An
output scaling factor is used when converting from signal intensity to
contrast agent concentration (see Perfusion
Analysis). This scaling factor will affect the actual pixel values in the
perfusion maps when deconvolution is applied. Typically this value should be
set so that the resulting perfusion and blood volume values in normal tissue are according to expected values. This
scaling factor must be specified since the exact relation between 1/T1, 1/T2
or 1/T2* and contrast agent concentration is not known in dynamic MRI
and depends on factors like contrast agent relaxivity, vascular structure,
tissue density and haematocrit. Note that even when AIF deconvolution is
applied pixel units are still in arbitrary units since the required
conversion factors are not assumed to be known. If the scaling factor is
correctly set according to calibration in normal subjects and known perfusion
/ blood volume values then the pixels values may be converted to mL/100 g and mL/100g/min respectively for CBV and CBF
images. In CT-perfusion analysis the pixel intensity is directly proportional
to contrast agent concentration and the scaling factor is then usually set to
a value of 1.
Advanced Options
Open
Advanced Options to access additional perfusion
settings.
Important note on SVD deconvolution:
It should be noted that the actual perfusion values (rBF, rMTT, Delay) obtained are
very dependent on the selected or determined SVD threshold, as shown in the
figure below. Although automatic (iterative) methods for threshold
determination are implemented in nordicICE, there is no guarantee that the
resulting perfusion values are clinically meaningful since many unknown
factors will influence the quality of the analysis.
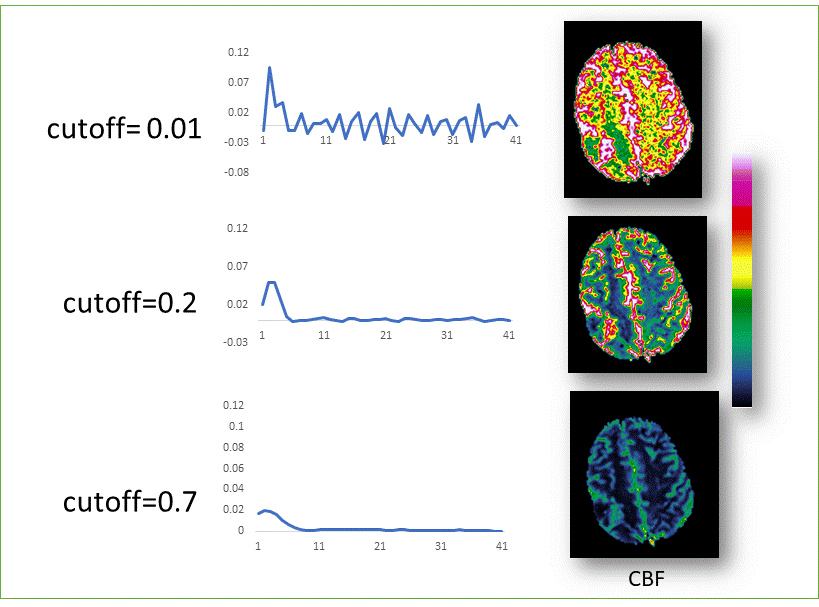
The
figure above shows the effect of varying the SVD threshold using the
‘truncated SVD’ option with manually defined SVD threshold. Both a too high cutoff (0.7, bottom CBF image) and a too low threshold (0.01,
top CBF image) result in erroneous perfusion maps. The middle row shows the
resulting residue functions for the same ROI in all three cases. The default
SVD cutoff used in the Perfusion Module (0.2) has
been set to provide reasonable perfusion estimates with the typical
signal-noise ratio obtained in a clinical situation (center images). There
is, however, no guarantee that this value will be optimal for all conditions.
Related topics:
Advanced Perfusion Options

|
|