T1
/ R1 relaxation analysis
Main
Menu: Modules->
This
module generates output images where the pixel intensities represents
longitudinal relaxation times or rates (T1/R1). The input images can be based
on one of the three following sequences:
- Inversion recovery (IR) with multiple (two or more)
inversion times (TIs).
- Saturation recovery with two or more delay times
(TDs)
- Spoiled gradient echo (SPGR) with two or more flip
angles (FAs)
Inversion (IR) - and saturation recovery (SR)
sequences:
IR/SR are sequences containing a 90 (SR) - or 180 deg
(IR) pre-pulse followed by images acquired at multiple delay (TD)
or inversion times (TI) either as a single shot or as multiple acquisitions:
The relaxation values are then obtained by performing a non-linear least
squares fit to the following expression:
SI = A(1
-K exp(-t*R1))
for
'real' images (i.e. when the pixel SI can take on
both positive and negative values) or:
SI = A | (1 -K exp(-t* R1)) | + C
for
modulus images (when negative signal intensities are converted to its modulus
value). In both cases, A and C are constants, K is a variable (equal to
2 for ideal inversion recovery and 1 for ideal saturation recovery), t
is the delay- (TD) or inversion time (TD/TI)
and R1 is the longitudinal relaxation rate (=1/T1). The constant C
accounts for the presence of a constant offset in the relaxation curve due to
the presence of noise so that the SI never reaches zero (only applicable for
modulus IR images)
Multiple flip
angle Spoiled Gradient Echo (SPGR) sequence:
For this sequence
type T1 or R1 are estimated based on spoiled gradient echo signal
expression:
SI =A sin(a)(1-exp(-TR/T1)/(1-cos(a)exp(-TR/T1)
where A is a constant, a is the flip angle (FA) and TR is the
repetition time
Two or more flip angles are needed and the resulting quality of the
relaxation maps depends strongly on the actual difference in SI for the
different FAs used. It is therefore important to choose FAs which gives
a large range of SIs for the T1-values of interest.
T1 Relaxation
analysis settings
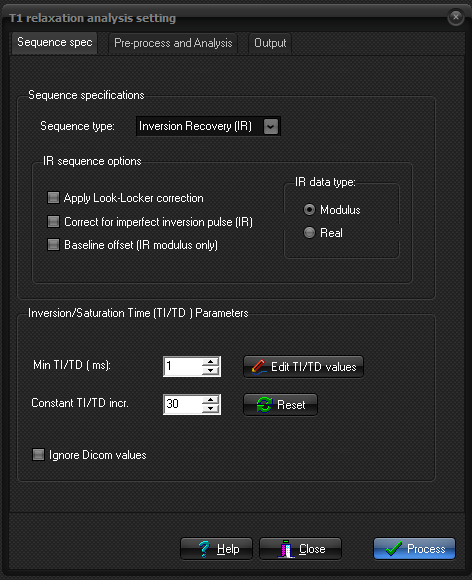
Sequence
specifications
Here
you specify which sequence (imaging method) is used to generate the data from
which T1 relaxation times will be calculated .
Three sequence types can be specified in the Sequence type drop-down
menu:
- Inversion Recovery (IR)
- Saturation recovery (SR)
- Multiple flip angle SPGR
If sequence type is set to either SR or
IR then multiple sequence related options can be set in the IR
sequence option panel:
- IR data type: This
can either be Modulus or Real. Se above for
details.
- Apply Look-Locker correction: Corrects the calculated T1/R1 values for the effect
of signal saturation in Look-Locker type sequences (multiple RF
excitation pulses after a single inversion pulse). The flip angle must
be specified when this option is enabled and
the option is only valid for IR curves.
- Correct for imperfect inversion pulse: When T1 is estimated from an Inversion Recovery (IR)
sequence, the formula used is: SI(TI)
= A[1- B exp(-TI/T1)] where TI
is the inversion time, A is a constant reflecting equilibrium
magnetization and B is a value reflecting the magnitude of the inversion
(180 deg) pulse. For a 'perfect' IR sequence
B=2, but the value may deviate from 2 when the B1-excitation field is
imperfect, causing the value of B to vary across the field of view. When
this option is enabled, the parameter B is allowed to vary (i.e. included as a parameter to be fitted). This will
account for variations in the inversion pulse, but also increases the
number of degrees of freedom in the fitting procedure, which can result
in greater variation in the resulting R1/T1 estimation. Note that when
the option 'Apply
Look-Locker correction' is enabled, correction for
imperfect inversion pulse is always applied, since this is part of the
Look-Locker correction algorithm.
- Baseline offset: When
enabled, the constant C in the signal
expression above is made a model parameter to account for the fact
that the signal level never reaches zero for modulus IR curves (due to
image noise). This option is not used for SR images or for real IR data.
Inversion
recovery and saturation recovery sequence parameters:
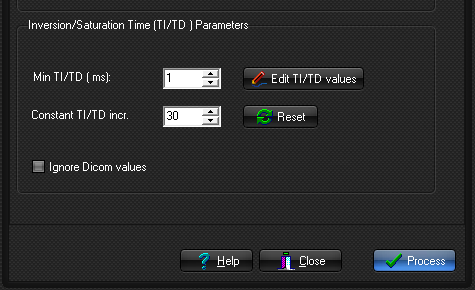
- Constant TI/TD spacing: Selected if a constant inversion or
delay time spacing is used. Min TI/TD is the time of
the first inversion/saturation time and Delta TI/TD
is the spacing of successive time-points after the first
TI/TD time. All times are in msec.
- Edit TI/TD values: The inversion-
or delay times are specified in a list. This would be used in
situations where the TI/TD values are
sampled at uneven intervals (and not specified in the DICOM header).
- Ignore Dicom values: If TI values are present in the DICOM header,
but one wishes to use other values.
Multiple
flip-angle SPGR sequence parameters:
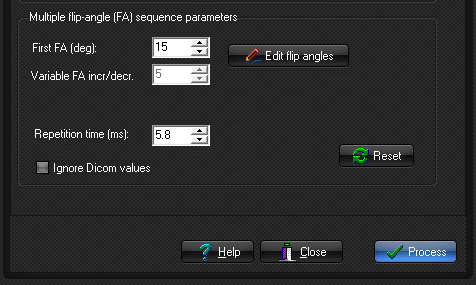
- Constant flip angle increment/decrement: Selected if a constant flip angle increment or
decrement is used. First FA is the flip angle of the first image and FA
increment/decrement is the constant change in flip angle for each
consecutive image in the series. All values are in degrees.
- Edit flip angles: The
flip angles used are specified in a list. This would be used in
situations where a non-constant flip angle increment or decrement is used
(and the values are not specified in the DICOM header).
- Repetition time:
Specifies the repetition time (TR) of the SPGR sequence (see sequence expression above).
- Ignore Dicom values: If valid flip angles and TR values are present
in the DICOM header, but one wishes to use other values.
Pre-process and
Analysis
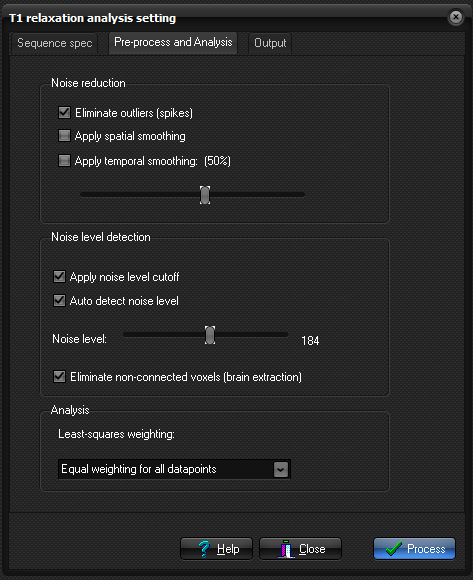
Noise reduction
options:
- Apply spatial smoothing: Apply a nearest neighbour smoothing kernel to
the raw data prior to curve fitting.
- Apply temporal smoothing: The dynamic time signal is smooted
(low-pass filtered), to reduce effects of noise
and spikes in the dynamic signal response. This smoothing does not
affect spatial resolution but may reduce the ability to detect rapid
signal changes. If temporal smoothing is selected the degree of
smoothing can be varied using the slider.
- Eliminate spikes:
Remove abnormally high (spiky) output values.
Noise Thresholding:
- Auto Detect: Automatically
determine the noise level in the input images (default). The noise level
can be modified using the slider.
- Show noise level cutoff: Show the selected noise threshold as red pixels in
the input images. All pixels colored red are
excluded from the analysis.
- Eliminate non-connected voxels (brain extraction): Removes areas that are not connected to the rest of
the brain.
Analysis:
It
is possible to use different weigthing for the
different datapoints. Variable
weighting can be useful in order to
apply less weight during the curve fitting procedure to data points known to
be affected by noise - or to ensure good curve fit for specific parts of the
curve. For instance, in an IR sequence, the data-points close to zero
magnetization values may be more prone to noise and could be given less weighting.
Or alternatively, very high amplitude data points can be given reduced
weighting to obtain better fits for data close to zero values.
Output
Here you specify options related to how the parametric images
are displayed.
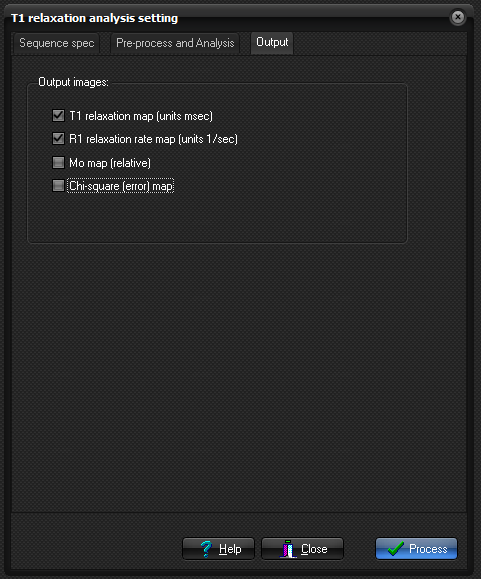
Output Image
types:
- R1 relaxation rate map: pixel values represent inverse relaxation times
(1/T1) in units of 1/sec.
- T1 relaxation rate map: pixels values represent relaxation times in units of
msec.
- M0 map:
This map represents the relative equilibrium magnetization obtained from
the curve fit. In the case of IR and SR sequences, this is obtained from
the intensity that is estimated for zero inversion- or saturation time.
For multi-flip angle sequences, the value of Mo represents an arbitrary
scaling factor needed to fit the equation for spoiled gradient echo
sequences to the measured signal variation.
- Chi-square map: An
additional output image is generated which represents the pixel-wise 'goodness of fit' (described by the
chi-square parameter resulting from the least squares fit)
Related topics:
T2
/ R2 relaxation analysis

|




