|
|
|
|
Dynamic Contrast
Enhanced (DCE) MRI analysis is the commonly used term to describe the
analysis of the transient effect of a contrast agent (CA) on the measured
signal intensity (or change in relaxation rate in the case of MRI) in the
tissue of interest following a rapid bolus injection of the CA. The effect of
the agent is analysed using an appropriate kinetic model, describing the
time-dependent distribution of the CA in tissue. The contrast agents
currently used in DCE analysis are most commonly small molecular weight
agents (like Gd -DTPA and similar chelates), which are renally excreted with
a half-life in blood limited by the glomerular filtration rate (GFR) of the
kidneys. The kinetics of these agents
in tissue can generally be described by a so-called two-compartment exchange
(TCx ) model (see e.g. Sourbron
et al Magn Reson Med,
2009 and Tofts et al. JMRI 1999 for details) . The two-compartment
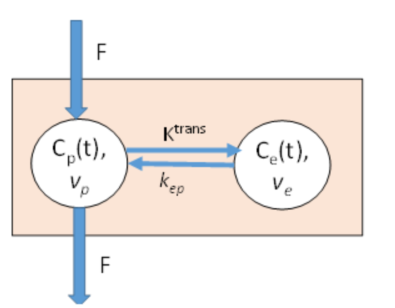
exchange model is illustrated in Figure1 below.
Figure
1. Two-compartment exchange (TCx
) model describing the capillary passage, and extravasation of contrast agent
(CA) in tissue. The CA enters the capillary volume due to tissue blood flow,
F, and is initially distributed in the tissue plasma volume, Vp , but is gradually equilibrated to the
extravascular, extracellular space (EES),
with volume fraction Ve at a rate
determined by the rate constants Ktrans
and kep,
where Ktrans =kep .
Ve . |
|
Although nordicICE provides analysis with the full TCx model, this model has four independent model parameters
(F, Vp, Ve,
Ktrans) and may give unstable results when
applied to pixel-wise dynamic MRI data if SNR is too low (Sourbron
and Buckley, Magn Reson
Med, 2011), and different simplifications and assumptions are therefore
commonly applied to obtain stable results, as described in more detail below.
The dynamic dose-response curve following a rapid injection of a
CA in a two-compartment system is typically described by a wash-in phase
(driven by tissue perfusion, F, and Ktrans ) followed by a wash-out phase (driven by
kep and plasma clearance). A
typical curve is shown in Figure 2 also
indicating some common descriptive metrics of the dynamic curve.
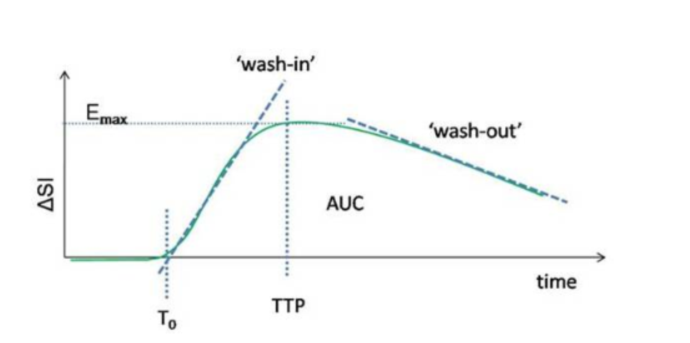
Figure
2. Typical dynamic
dose-response following rapid contrast injection in a
two-compartment exchange system, and corresponding curve
parameters. AUC is the area under the dynamic response curve and TTP is
the time to peak enhancement.
In case of very slow
leakage, large EES or too short total sampling time, the wash-out phase can be
ill defined. Also, the wash-in phase can often be bi-phasic with an initial
steep slope (perfusion driven) followed by a reduced wash-in rate (Ktrans driven).
The descriptive
parameters described above do not directly reflect the underlying parameters of
the kinetic model described in Figure 1. To extract these tissue specific
kinetic parameters, the tissue response curve, and the arterial input function
(AIF, describing the dynamic concentration time curve of the bolus passage in
an artery feeding the tissue of interest) must be used as input to an
appropriate mathematical model of the CA kinetics. As mentioned above, the full
TCx model is complex, and simplifications are
typically applied to obtain robust results when attempting to fit the
pixel-wise dynamic data to the model.
The kinetic models
available in nordicICE are summarized below.
1. Two-compartment
exchange (TCx) model
This is the complete
four-parameter model as shown in Figure 1, including both perfusion (F), plasma
volume (Vp), Ktrans
and extracellular extravascular volume (Ve).
The full model is mathematically complex, and the resulting impulse response is
a bi-exponential function of the four model parameters. In nordicICE, the model
is estimated by matrix inversion, as described in: Flouri
et al Magn Reson Med (2016)
2.
Extended Tofts (ET) model.
This model assumes rapid flow
(perfusion) relative to the sampling rate of the MRI sequence so that the CA
transit time through tissue is essentially instantaneous. Under these
conditions, it can be shown that the total CA in tissue is given by;
Eq
1
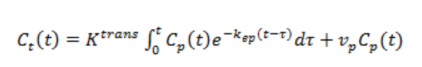
where Cp is the CA concentration in plasma and Ct is
the total tissue CA concentration. Eq. 1 is also-called convolution integral,
describing the CA kinetics (CA concentration change) in tissue following a
bolus of CA initially present in the arterials feeding the tissue. In DCE-MRI,
Cp (t) is determined from a well-defined artery (arterial input
function, AIF) feeding the tissue of interest.
3. Tofts-Kermode (TK) model
This is a simplification of the ET model where the plasma volume is assumed to
be negligible so that:
Eq 2
![]()
4. Patlak model
This model assumes the back-flux of CA from EES to plasma space to be
negligible during the measurement time (i.e. exp ( -kep
T ) ⁓ 1, where
T=total measurement time). The kinetic model then becomes:
Eq 3
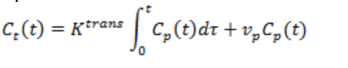
The advantage of the Patlak model is that is can be linearized, e.g. the
expression can be re-cast in to a linear form so that Ktrans
and vp can
be determined by linear regression analysis (compared to non-linear regression
for Eqs 1,2), which is generally more stable and less
sensitive to image noise. The Patlak model can
therefore be a good alternative if leakage rates are known to be low or when
the DCE-MRI data has low SNR, giving poor curve fits for the non-linear models.
5.
Incremental model
This method iterates through multiple
models with increasing complexity and number of model parameters (Bagher-Ebadian et al, Magn Reson Med, 2012). For each tested model, the
goodness-of-fit of the model to the data is compared to the fit of previous
(less complex) model and a statistical test (F-test) is performed to test if
the fit is significantly better than for the previous (less complex) model,
justifying the increased complexity. The following models are included in the
incremental testing:
a. Noise
only (zero Vp , Ktrans and kep
) -> Pixels shown as black in model selection map
b.
No leakage (Non-zero Vp ;
zero Ktrans and kep ) -> blue pixels
c.
Patlak model (non-zero Ktrans
and Vp , zero kep ) -> cyan pixels
d.
Tofts-Kermode model (non-zero Ktrans and
kep , zero Vp
) -> yellow pixels
e.
Extended Tofts model (non-zero Vp , Ktrans and kep
) -> green pixels
f.
Two-compartment exchange (TCx) model (F, Vp, Ve and Ktrans)
-> red pixels
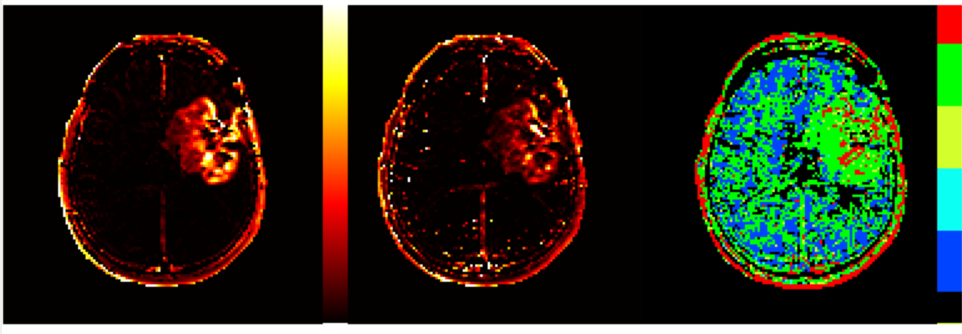
Figure
3 Sample
Ktrans parametric maps obtained
with Two-compartment exchange (TCx) model and Patlak model. Note that use of Patlak
model here provides less noise in the resulting map. Despite the Patlak based model map appearing less noisy, the model
selection map (obtained by using the “incremental modelling” option) indicates
that best fit in tumor region is obtained using
either ET model (green) or TCx model (red) models.
Note that in the model selection map palette has six distinct colors - one of each model to be tested, as described
above.
Colour map for incremental model:
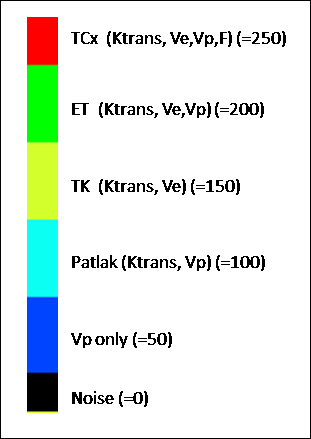
Kinetic
modelling with vascular deconvolution
If the arterial input function
(AIF) can be measured then the equation describing the selected kinetic model (Eqs . 1-3) can be solved using standard
numerical methods. In nordicICE, the AIF can be determined from many different
methods, ranging from manual detection from user selected ROI, global automatic
detection, or region specific automatic detection and finally using pre-defined
(population) AIFs. Figure 4 shows a sample AIF, tumor tissue curve and corresponding curve fit. The AIF
contains signal from whole blood whereas the kinetic model assumes that we
measure the plasma concentration of contrast agent. We therefore also need to
scale the AIF signal according to the blood hematocrit
(Hct ) since CAIF = (1-Hct)Cp .The
Hct can be specified in nordicICE but is by default
set to 0.45.
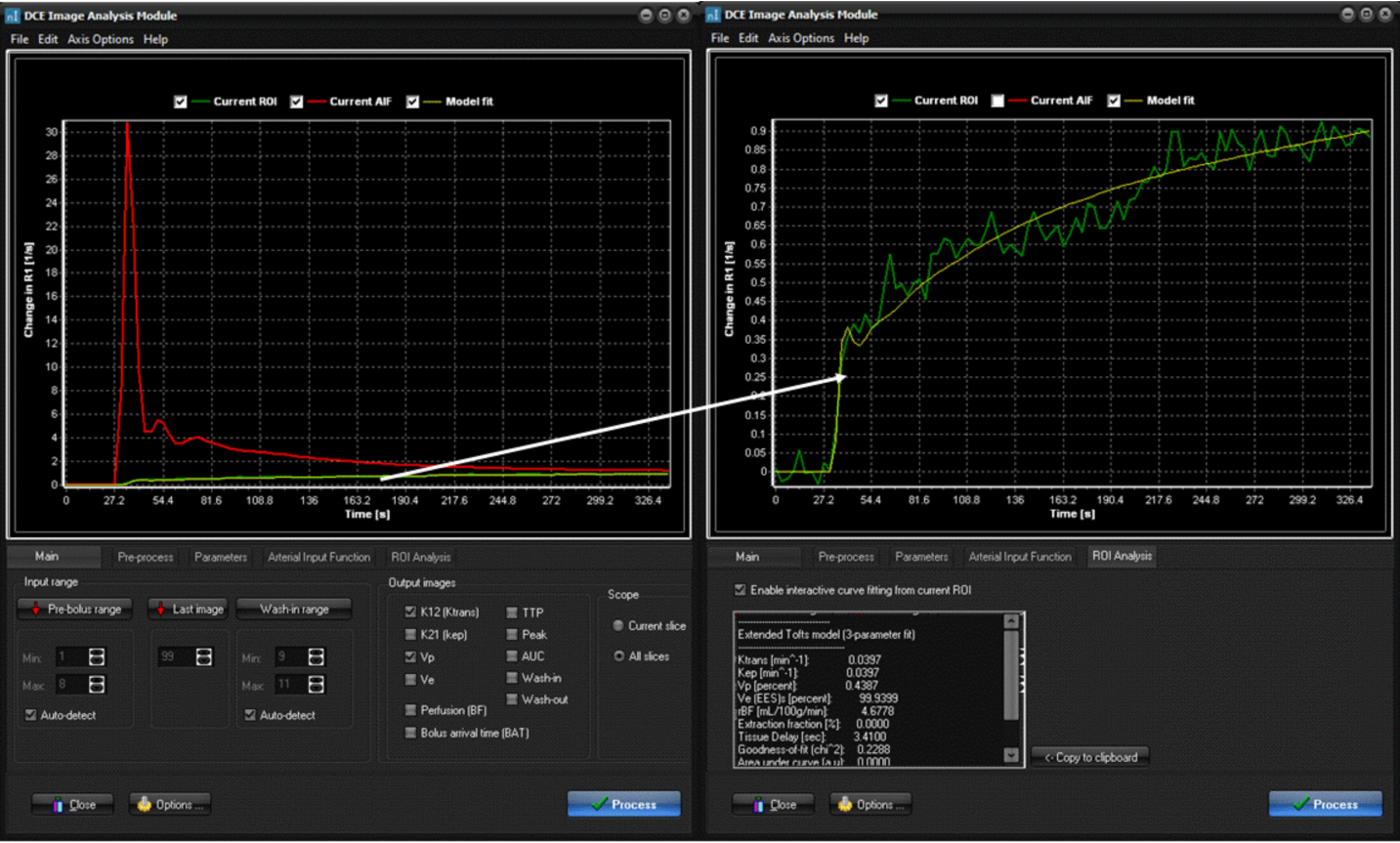
Figure
4. AIF
determination and model fit visualization in nordicICE. Here, the AIF (red
curve) was determined using the “automatic detection” option. The right panel
shows the interactive curve fit option where the curve fit (yellow line) of a
ROI in a tumor region (green curve) is shown, using
the “ incremental model” selection
option. The kinetic parameters obtained from the curve fit is shown in the
panel below the curve.
Estimation of concentration time curves (CTC)
The kinetic models
used to MRI-DCE analysis explicitly assumes that the CA concentration (C) is
known. In MRI, the CA concentration cannot be measured directly and has to be
derived from the observed change in MR signal intensity (SI) in response to the
presence of the CA in tissue. One common assumption is that the change in 1/T1
relaxation rate is proportional to C, which is valid as long as there is a fast
water exchange between water protons in tissue and the paramagnetic center of the CA. Given this assumption, C can be estimated
(in relative units) given the change in 1/T1 relaxation rate can be measured.
From strongly T1-weighted sequences, it is often assumed that the change 1/T1
is proportional to the change in SI. From this assumption, relative change in
1/T1 (and hence C) can then be estimated simply from the relative change
in SI (either in absolute units or as a percent change). Both these conversion
options are available in the DCE module and are typically used if the exact
details of the MR sequence used are not known or a sequence is used where the
signal response cannot be expressed in a simple analytical form (e.g. non
steady-state sequences). The DCE module also enables exact estimation of change
in 1/T1 for specific MR sequences. The two types of sequences which can
currently be modelled are:
· Spoiled gradient echo (GRE) sequences. This is the most common sequence used in DCE imaging and is a
class of sequences where the SI is given by the following expression:
Eq. 4
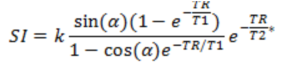
where k is an
(unknown) constant, α is the flip angle, TR is the repetition time, T1 and
T2* are the relaxation times and TE is the echo time.
· Saturation recovery (SR ) This is an
alternative to the standard spoiled GRE sequence where the DCE signal is read
out following given delay after a 90 degree preparation pulse. Here, the SI is
given by :
Eq. 5
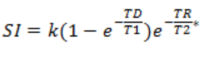
where TD is the delay time (time from 90-degree pulse to start of
acquisition). This class of sequence has been shown to be less sensitive to
water exchange effects when short TD-times are used (Larsson et al. Magn Reson Med, 2001).
By
assuming the TE<<T2*, the change in 1/T1 can then be determined if TR and
flip angle (GRE) or TD (SR)are known, in addition to 1/T1 at baseline (1/T10).
In the DCE module 1/T10 can either be set at a fixed values (1000 ms as default) or can be taken from a T1-relaxation map
estimated separately (either using the nordicICE
T1-relaxation module or by separate software) . For MR sequences which
cannot be modelled by any of the two expressions, the relative change in 1/T1
must be estimated from the relative (or absolute) change in SI, which then
assumes that this SI is linearly related to 1/T1 changes induced by the CA in
tissue and blood.